
Hatasu Kobayashi 1, Yurie Mori 2,3, Sharif Ahmed 4, Yuichiro Hirao 5,6, Shinya Kato 7, Shosuke Kawanishi 8, Mariko Murata 9, Shinji Oikawa 10,✉,2
- Author information
- Article notes
- Copyright and License information
PMCID: PMC10132762 PMID: 36461940
Abstract
Molnupiravir is an antiviral agent recently used for treating coronavirus disease 2019 (COVID-19). Here, we demonstrate that N4-hydroxycytidine (NHC), a molnupiravir metabolite, treated with cytidine deaminase (CDA) induced Cu(II)-mediated oxidative DNA damage in isolated DNA. A colorimetric assay revealed hydroxylamine generation from CDA-treated NHC. The site specificity of DNA damage also suggested involvement of hydroxylamine in the damage. Furthermore, Cu(I) and H2O2 play an important role in the DNA damage. We propose oxidative DNA damage via CDA-mediated metabolism as a possible mutagenic mechanism of NHC, highlighting the need for careful risk assessment of molnupiravir use in therapies for viral diseases, including COVID-19.
Keywords: 8-oxo-7, 8-dihydro-2′-deoxyguanosine, DNA damage, cytidine deaminase, hydroxylamine, molnupiravir, N 4-hydroxycytidine, reactive oxygen species
N 4-hydroxycytidine (NHC) is an active metabolite of the SARS-CoV-2 antiviral molnupiravir. NHC treated with cytidine deaminase induced Cu(II)-mediated generation of reactive oxygen species and DNA damage, which is proposed as a new mutagenic mechanism of NHC.
Molnupiravir (EIDD-2801), an oral antiviral agent, has been approved, or has been filed for approval, for use in the treatment of coronavirus disease 2019 (COVID-19) in multiple countries. Molnupiravir is an isopropyl ester prodrug of the antiviral ribonucleoside analogue N4-hydroxycytidine (NHC; EIDD-1931) [1]. In plasma, molnupiravir is cleaved by esterase to release NHC, which leads to mutations in the viral RNA genome of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) [1]. The accumulation of mutations impairs faithful genome replication by the virus and prevents viral propagation.
The Indian Council of Medical Research excluded molnupiravir from its COVID-19 treatment guidelines [2], revealing that there remain concerns regarding the safety of molnupiravir. Zhou et al reported that NHC induced mutations in cultured mammalian cells [3], raising the concern that molnupiravir, which is converted into NHC, is potentially mutagenic in humans [4]. Interestingly, a recent study showed that biomarkers of oxidative stress were upregulated in cultured cells treated with NHC [5]. Thus, there is a possibility that reactive oxygen species (ROS) are involved in the mutagenicity of NHC, but the mechanisms remain unknown.
Previously, we reported that acetamide and acrylohydroxamic acid, which, like NHC, possess hydroxylamine groups, cause ROS generation and oxidative DNA damage through the enzymatic generation of hydroxylamine (NH2OH) in the presence of Cu(II) [6, 7]. Therefore, in the present study, to clarify the mutagenic mechanism of NHC, we investigated the DNA damage caused by NHC treated with cytidine deaminase (CDA), which is expected to metabolize NHC and to generate hydroxylamine, in the presence of Cu(II). It is known that a substantial amount of copper is located in the cell nucleus and that much of it is bound to DNA bases [8]. Copper ions possess properties that aid DNA damage by other agents and enhance mutagenic activities via the production of ROS [8]. The content of 8-oxo-7,8-dihydro-2′-deoxyguanosine (8-oxodG), which is a relevant indicator of oxidative DNA damage and mutagenic lesions [9], in calf thymus DNA was measured using a high-performance liquid chromatograph equipped with an electrochemical detector (HPLC-ECD). We also evaluated the generation of hydroxylamine from CDA-treated NHC by a colorimetric assay. Furthermore, we investigated the effects of ROS scavengers on the DNA damage and the site specificity of the DNA damage using a 32P-5′-end-labelled DNA fragment of a human cancer-related gene.
METHODS
Analysis of 8-oxodG Formation in Calf Thymus DNA
Measurement of 8-oxodG in calf thymus DNA was performed as described previously [9]. After incubation of NHC (Sigma-Aldrich) with 125 µg/mL CDA (human, recombinant; Sigma-Aldrich) for 16 hours at 37°C, the reaction mixtures, containing 100 µM/base calf thymus DNA, CDA-treated NHC, and 100 µM CuCl2 in 400 µL of 4 mM sodium phosphate buffer (pH 7.8) containing 5 µM diethylenetriamine-N, N, N′, N′′, N′′-pentaacetic acid (DTPA), were incubated for 1 hour at 37°C. After ethanol precipitation, the DNA was digested into nucleosides with nuclease P1 and calf intestine phosphatase. The amount of 8-oxodG was measured using an HPLC system (SLC-10Avp, LC-20AD, SPD-10AVvp; Shimadzu Corp) equipped with an ECD (HTEC-510; Eicom).
Spectrophotometric Determination of Hydroxylamine
As in a previous report [10], hydroxylamine evaluation was performed in two steps: iodine-mediated oxidation of hydroxylamine to nitrite, followed by spectrophotometric determination of nitrite using N-(1-naphthyl)ethylenediamine. After incubation of NHC with 125 µg/mL CDA for 16 hours at 37°C, the reaction mixtures containing CDA-treated NHC, 140 µM iodine, and 750 µM sulfanilic acid in 400 µL of 2.5% acetic acid were incubated for 30 minutes at 37°C, and then excess iodine was removed by solid-phase extraction on a Sep-Pak C18 cartridge (Waters Corp). After adding N-(1-naphthyl)ethylenediamine dihydrochloride (75 µM), the absorbance was measured at 540 nm using a microplate reader (Bio-Rad Laboratories).
Detection of Damage to Isolated 32P-labelled DNA Fragments
A fragment containing exon 2 of the human p16 tumor suppressor gene was obtained as described previously [11]. The 5′ end-labelled 460-base pair fragment (EcoRI* 9481–EcoRI* 9940) was further digested with BssHII to obtain the singly labelled 309-base pair fragment (EcoRI* 9481–BssHII 9789) and the 147-base pair fragment (BssHII 9794–EcoRI* 9940). An asterisk indicates 32P labelling.
NHC was treated with 125 µg/mL CDA for 16 hours at 37°C and then was added to reaction mixtures containing the 32P-labelled DNA fragment, 20 µM/base calf thymus DNA, and 100 µM CuCl2 in 200 µL of 10 mM sodium phosphate buffer (pH 7.8) containing 5 µM DTPA. When examining the effects of bathocuproine and ROS scavengers such as ethanol, mannitol, sodium formate, 3-(methylthio) propionaldehyde (methional), catalase, and superoxide dismutase (SOD), these reagents were added before the addition of CDA-treated NHC. After incubation for 1 hour at 37°C, the DNA fragments were heated in 1 M piperidine for 20 minutes at 90°C, followed by electrophoresis on an 8% polyacrylamide/8 M urea gel. An autoradiogram was obtained by exposing an X-ray film (Fujifilm Corp) to the gel.
The preferred cleavage sites were determined by directly comparing the positions of the oligonucleotides with those produced by the chemical reactions of the Maxam-Gilbert procedure, using a DNA-sequencing system (LKB 2010 Macrophor; LKB Pharmacia Biotechnology Inc.). The autoradiogram was obtained by exposing an imaging plate (BAS-MS2040; Fujifilm Corp) to the gel. The relative amounts of oligonucleotides from the treated DNA fragments were measured using a laser scanner (Typhoon FLA-9500; GE Healthcare) and analyzed with ImageQuant TL software (GE Healthcare).
RESULTS
Formation of 8-oxodG in Calf Thymus DNA by CDA-Treated NHC in the Presence of Cu(II)
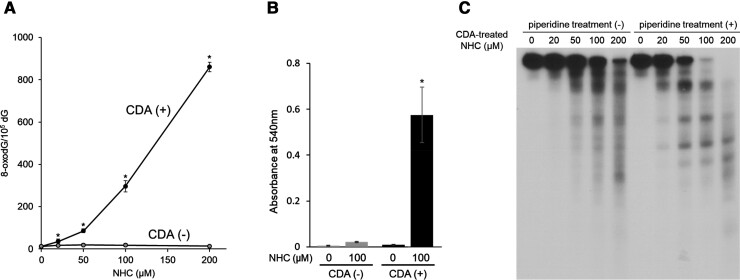
We measured the content of 8-oxodG in calf thymus DNA following incubation with NHC, treated with or without CDA, in the presence of Cu(II). The level of 8-oxodG was significantly increased by CDA-treated NHC in a concentration-dependent manner, whereas NHC without CDA treatment did not induce the formation of 8-oxodG (Figure 1A), indicating that CDA treatment is essential for NHC-mediated 8-oxodG formation.
Generation of Hydroxylamine From NHC Treated With CDA
To clarify whether CDA treatment leads to the generation of hydroxylamine from NHC, we performed a colorimetric assay to evaluate hydroxylamine generation from NHC treated with or without CDA. The absorbance at 540 nm was significantly increased for NHC treated with CDA compared with untreated NHC (Figure 1B), indicating that hydroxylamine was generated from NHC via CDA-mediated metabolism.
Damage of 32P-labelled DNA Fragments by CDA-Treated NHC Plus Cu(II)
Figure 1 C shows the autoradiogram of DNA fragments incubated with CDA-treated NHC in the presence of Cu(II), treated with or without piperidine. Oligonucleotides were detected on the autoradiogram as a result of the DNA cleavage. CDA-treated NHC plus Cu(II) caused DNA cleavage in a concentration-dependent manner. Furthermore, the DNA cleavage was enhanced by piperidine treatment. As piperidine cleaves DNA at sugars with modified bases, these results suggest that CDA-treated NHC caused base modification as well as breakage of the deoxyribose phosphate backbone in the presence of Cu(II).
Effects of ROS Scavengers and a Cu(I) Chelator on the DNA Damage Induced by CDA-Treated NHC
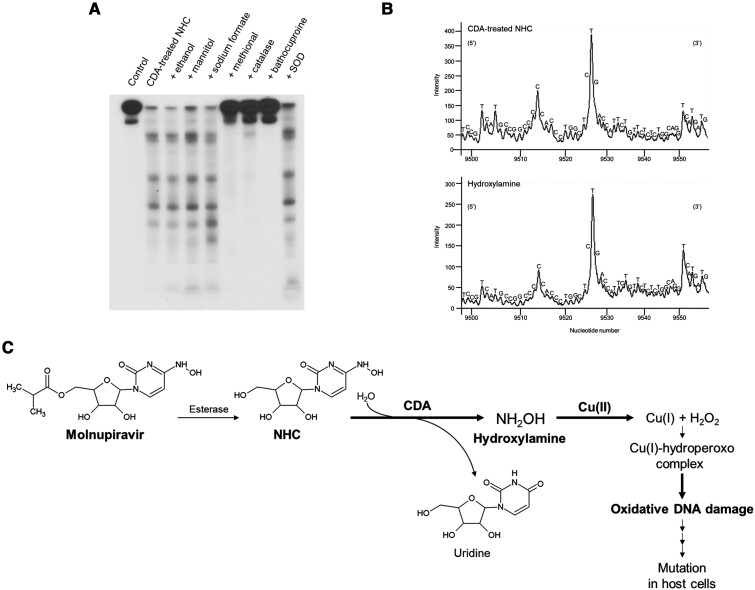
We examined the effects of various ROS scavengers and bathocuproine on the damage of 32P-labelled DNA fragments by CDA-treated NHC plus Cu(II). While typical free hydroxyl radical (•OH) scavengers (ethanol, mannitol, and sodium formate) did not inhibit DNA damage, methional, which scavenges ROS with weaker reactivity than •OH such as the Cu(I)-hydroperoxo complex, did prevent DNA damage (Figure 2A). Catalase, an H2O2 scavenger, and bathocuproine, a Cu(I)-specific chelator, also showed inhibitory effects on the DNA damage (Figure 2A). The DNA damage was not prevented by SOD (Figure 2A), which rapidly catalyzes the dismutation of superoxide anion radical (O2•−) to H2O2 in vivo. These results suggest that the Cu(I)-hydroperoxo complex, which is produced from H2O2 and Cu(I), is essential for the DNA damage induced by CDA-treated NHC in the presence of Cu(II).
Site Specificity of the DNA Damage Induced by CDA-Treated NHC
The site specificity of DNA damage induced by CDA-treated NHC in the presence of Cu(II) was determined by DNA sequencing using the Maxam-Gilbert procedure. The relative intensity of DNA damage obtained by scanning an autoradiogram with a laser scanner is shown in Figure 2B. CDA-treated NHC plus Cu(II) caused piperidine-labile lesions frequently at thymine (T) and at some cytosine (C) residues in DNA fragments obtained from the p16 tumor suppressor gene (Figure 2B, upper). We compared the site specificity of the DNA damage induced by CDA-treated NHC with that induced by hydroxylamine. The site specificity of DNA damage caused by hydroxylamine plus Cu(II) (Figure 2B, lower) was almost the same as that caused by CDA-treated NHC.
DISCUSSION
In the present study, we demonstrated that, in the presence of Cu(II), CDA-treated NHC increased the formation of 8-oxodG, a potentially mutagenic oxidative form of DNA base damage [9], in calf thymus DNA, whereas NHC without CDA treatment did not. The DNA damage induced by CDA-treated NHC plus Cu(II) was confirmed using 32P-labelled human DNA fragments obtained from the p16 tumor suppressor gene. An assessment of the site specificity of the oxidative DNA damage revealed that CDA-treated NHC plus Cu(II) induced piperidine-labile sites at T and C residues. Interestingly, the site specificities of the Cu(II)-mediated oxidative DNA damage induced by CDA-treated NHC and by hydroxylamine were almost identical. In addition, hydroxylamine generation in NHC with CDA treatment was confirmed by a colorimetric assay. These results suggest that hydroxylamine generated from NHC treated with CDA induces oxidative DNA damage. CDA, a ubiquitous enzyme that plays a major role in recycling free pyrimidines, deaminates cytidines [12]. CDA is known to catalyze a variety of synthetic nucleoside analogues in addition to its natural substrates [12]. It is therefore possible that CDA catalyzes NHC with a structure similar to that of cytidine. Collectively, CDA is a critical mediator for NHC-induced oxidative DNA damage in the presence of Cu(II).
To clarify the types of ROS involved in the oxidative DNA damage, we examined the effects of ROS scavengers and a Cu(I) chelator on the oxidative DNA damage using 32P-labelled DNA fragments. The results showed that the oxidative DNA damage was inhibited by a Cu(I) chelator and scavengers of H2O2 and a variety of ROS other than •OH, whereas it was not inhibited by typical •OH scavengers. This suggests that Cu(I)-hydroperoxo complex generated from Cu(I) and H2O2 plays an important role in the oxidative DNA damage. From these results and our previous study [7], we propose a possible mechanism, shown in Figure 2C, in which NHC treated with CDA oxidatively damages DNA. When NHC, which is converted from molnupiravir, is metabolized by CDA, hydroxylamine is generated. This generated hydroxylamine then induces Cu(II)-mediated oxidative DNA damage, possibly through Cu(I)-hydroperoxo complex generation. These results indicate that it is important to focus on the processing of NHC by CDA when investigating the mutagenicity of NHC.
Because the triphosphate form of NHC is incorporated into viral RNA as a substrate and induces mutation [13, 14], it has generally been assumed that NHC may cause mutation in the host genome through the incorporation of NHC-triphosphate and N4-hydroxy-deoxycytidine triphosphate into host DNA [3]. In the present study, we propose oxidative DNA damage resulting from CDA-mediated hydroxylamine generation from NHC as another possible mechanism underlying the mutagenicity of NHC. The maximum blood concentration of NHC reached approximately 14 μM (3640 ng/mL) after a single dose of 800 mg of molnupiravir in humans [15]. In this study, under CDA treatment, NHC induced DNA damage at 20 μM. CDA has been reported to metabolize various nucleoside analogues in humans [12]. Thus, there is a possibility that hydroxylamine produced from NHC could cause DNA damage in vivo. Future studies are warranted to investigate the level of hydroxylamine in animals and humans dosed with molnupiravir. Finally, our results and these previous reports highlight the need for a careful risk assessment of molnupiravir use in therapies for viral infections including COVID-19.
Contributor Information
Hatasu Kobayashi, Department of Environmental and Molecular Medicine, Mie University Graduate School of Medicine, Tsu, Japan.
Yurie Mori, Department of Environmental and Molecular Medicine, Mie University Graduate School of Medicine, Tsu, Japan; Faculty of Pharmacy, Gifu University of Medical Science, Kani, Japan.
Sharif Ahmed, Department of Environmental and Molecular Medicine, Mie University Graduate School of Medicine, Tsu, Japan.
Yuichiro Hirao, Department of Environmental and Molecular Medicine, Mie University Graduate School of Medicine, Tsu, Japan; Mie Prefectural College of Nursing, Tsu, Japan.
Shinya Kato, Radioisotope Experimental Facility, Advanced Science Research Promotion Center, Mie University, Tsu, Japan.
Shosuke Kawanishi, Faculty of Pharmaceutical Science, Suzuka University of Medical Science, Suzuka, Japan.
Mariko Murata, Department of Environmental and Molecular Medicine, Mie University Graduate School of Medicine, Tsu, Japan.
Shinji Oikawa, Department of Environmental and Molecular Medicine, Mie University Graduate School of Medicine, Tsu, Japan.
Notes
Financial support. This work was supported by the Japan Society for the Promotion of Science KAKENHI (grant numbers 19H03885 and 18K19673). Funding to pay the Open Access publication charges for this article was provided by the Japan Society for the Promotion of Science.
References
- 1. Tian L, Pang Z, Li M, et al. Molnupiravir and its antiviral activity against COVID-19. Front Immunol 2022; 13:855496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Extance A. Covid-19: what is the evidence for the antiviral molnupiravir? BMJ 2022; 377:o926. [DOI] [PubMed] [Google Scholar]
- 3. Zhou S, Hill CS, Sarkar S, et al. β-D-N4-hydroxycytidine inhibits SARS-CoV-2 through lethal mutagenesis but is also mutagenic to mammalian cells. J Infect Dis 2021; 224:415–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. van Schalkwyk JM. Buyer beware: molnupiravir may damage DNA. BMJ 2021; 375:n2663. [DOI] [PubMed] [Google Scholar]
- 5. Brandsma I, Derr R, Zhang G, Moelijker N, Hendriks G, Østerlund T. Genotoxicity assessment of potentially mutagenic nucleoside analogues using ToxTracker®. Toxicol Lett 2022; 362:50–8. [DOI] [PubMed] [Google Scholar]
- 6. Sakano K, Oikawa S, Hiraku Y, Kawanishi S. Mechanism of metal-mediated DNA damage induced by a metabolite of carcinogenic acetamide. Chem Biol Interact 2004; 149:52–9. [DOI] [PubMed] [Google Scholar]
- 7. Mori Y, Kobayashi H, Fujita Y, et al. Mechanism of reactive oxygen species generation and oxidative DNA damage induced by acrylohydroxamic acid, a putative metabolite of acrylamide. Mutat Res Genet Toxicol Environ Mutagen 2022; 873:503420. [DOI] [PubMed] [Google Scholar]
- 8. Linder MC. The relationship of copper to DNA damage and damage prevention in humans. Mutat Res 2012; 733:83–91. [DOI] [PubMed] [Google Scholar]
- 9. Kasai H, Crain PF, Kuchino Y, Nishimura S, Ootsuyama A, Tanooka H. Formation of 8-hydroxyguanine moiety in cellular DNA by agents producing oxygen radicals and evidence for its repair. Carcinogenesis 1986; 7:1849–51. [DOI] [PubMed] [Google Scholar]
- 10. Verma P, Gupta VK. A sensitive spectrophotometric method for the determination of hydroxylamine by use of p-nitroaniline and N-(1-naphthyl)-ethylenediamine. Talanta 1984; 31:1013–4. [DOI] [PubMed] [Google Scholar]
- 11. Kobayashi H, Mori Y, Iwasa R, et al. Copper-mediated DNA damage caused by purpurin, a natural anthraquinone. Genes Environ 2022; 44:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Serdjebi C, Milano G, Ciccolini J. Role of cytidine deaminase in toxicity and efficacy of nucleosidic analogs. Expert Opin Drug Metab Toxicol 2015; 11:665–72. [DOI] [PubMed] [Google Scholar]
- 13. Kabinger F, Stiller C, Schmitzová J, et al. Mechanism of molnupiravir-induced SARS-CoV-2 mutagenesis. Nat Struct Mol Biol 2021; 28:740–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gordon CJ, Tchesnokov EP, Schinazi RF, Götte M. Molnupiravir promotes SARS-CoV-2 mutagenesis via the RNA template. J Biol Chem 2021; 297:100770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Painter WP, Holman W, Bush JA, et al. Human safety, tolerability, and pharmacokinetics of molnupiravir, a novel broad-spectrum oral antiviral agent with activity against SARS-CoV-2. Antimicrob Agents Chemother 2021; 65:e02428-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
Articles from The Journal of Infectious Diseases are provided here courtesy of Oxford University Press
